Diels Alder Reaction
A frequent experiment in computational chemistry is mechanism elucidation which requires reaction path searches. Of course the here presented example is so small that a DFT path search would easily be doable but for the sake of simplicity we nevertheless go for a plain Diels-Alder reaction at the GFN2-xTB (gas) level.
Optional Exercise: Building the Structures
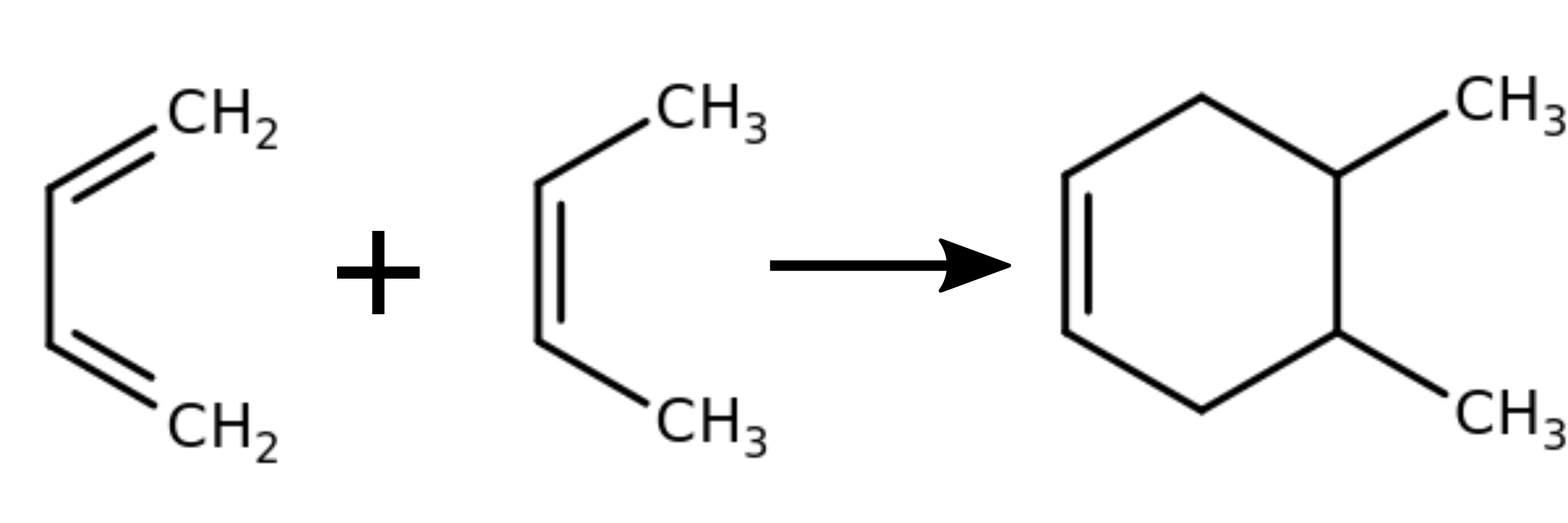
Build the input structures (buta-1,3-diene and but-2-ene) yourself with a molecular editor, e.g. avogadro. Pay attention that the atom order is the same in the start and end structure.
Since the construction can be cumbersome, we also provide the start and end structures below.
22
C -1.28276 2.69140 0.40114
C -0.10622 3.47437 -0.10487
C -1.31725 1.34886 0.39739
C -0.18963 0.50818 -0.12814
H 0.09446 3.22837 -1.16875
H 0.79145 3.24332 0.50544
H -0.32128 4.56048 -0.02803
H 0.73638 0.70010 0.45170
H -0.01088 0.73766 -1.20004
H -0.45423 -0.56621 -0.03962
C -4.15786 2.74293 -0.82082
C -4.13449 1.25635 -0.75405
C -3.15708 3.50005 -1.28913
C -3.11517 0.49310 -1.17571
H -2.21537 0.91108 -1.60550
H -3.18122 -0.58578 -1.08070
H -3.26017 4.58033 -1.29328
H -2.23255 3.07727 -1.66093
H -5.05032 3.24434 -0.45751
H -5.00213 0.76050 -0.33223
H -2.14406 3.23510 0.77856
H -2.20376 0.84820 0.77610
22
C -1.60263 2.81026 -0.14343
C -0.19808 3.43006 -0.17459
C -1.64659 1.22316 -0.03985
C -0.31094 0.50969 -0.31334
H 0.35069 3.11556 -1.08734
H 0.37757 3.13476 0.72718
H -0.27170 4.53852 -0.17884
H 0.46981 0.84157 0.40135
H 0.03500 0.70341 -1.35082
H -0.43141 -0.58687 -0.17934
C -3.85714 2.76279 -1.12879
C -3.97924 1.44105 -0.94234
C -2.47150 3.33574 -1.31197
C -2.72876 0.60996 -0.97523
H -2.35337 0.65325 -2.02228
H -2.93209 -0.45611 -0.73712
H -2.52163 4.44604 -1.32265
H -2.04129 2.99420 -2.27859
H -4.72692 3.41029 -1.07982
H -4.94744 0.99186 -0.74611
H -2.09637 3.20130 0.77678
H -1.92079 0.98738 1.01317
Pathfinder
Perform a reaction path search at the GFN2-xTB (gas) level of theory for the given reactants. You need the following input file (path.inp) in addition to your coordinate files:
xtb start.xyz --path end.xyz --input path.inp > path.out &
$path
nrun=1 # number of runs
npoint=25 # number of points on path
anopt=10 # how many optimization steps per point
kpush=0.003 # push factor for path search
kpull=-0.015 # pull factor for path search
ppull=0.05
alp=1.2
$end
The resulting path should be written to xtbpath.xyz.
Growing String Method
Another option for reaction path search is the growing string method (GSM) developed by the Zimmermann group and adapted for xtb. To set it up, you’ll need inpfileq (provided), ograd (provided) which you can download here.
First, unpack the downloaded archive and place the gsm.orca program in your path.
Next, go to your working directory and place inpfileq and ograd there.
Then, you have to create a direcotry named scratch and move the tm2orca.py script into it.
Concatenate the start.xyz and end.xyz file into initial0000.xyz and move the latter also into the scratch directory.
Finally, make the ograd and tm2orca.py scripts are executable with
chmod u+x ograd
chmod u+x scratch/tm2orca.py
Start the calculation with the following command or use the run.sh script to check if you followed all the above steps and all files are in the correct location.
gsm.orca > gsm.out 2> end &
#!/bin/bash
if ! command -v gsm.orca >/dev/null 2>&1; then
echo "The program 'gsm.orca' was not found."
exit 1
fi
if ! command -v xtb >/dev/null 2>&1; then
echo "The program 'xtb' was not found."
exit 1
fi
if [[ ! -f "ograd" ]]; then
echo "The file 'ograd' was not found."
exit 1
fi
if [[ ! -x "ograd" ]]; then
echo "The file 'ograd' is not executable."
exit 1
fi
if [[ ! -d "scratch" ]]; then
echo "The directory 'scratch' was not found."
exit 1
fi
if [[ ! -f "scratch/initial0000.xyz" ]]; then
echo "The file 'scratch/initial0000.xyz' was not found."
echo "You can create it with the following command:"
echo "cat start.xyz end.xyz > scratch/initial0000.xyz"
exit 1
fi
if [[ ! -f "scratch/tm2orca.py" ]]; then
echo "The file 'scratch/tm2orca.py' was not found."
exit 1
fi
if [[ ! -x "scratch/tm2orca.py" ]]; then
echo "The file 'scratch/tm2orca.py' is not executable."
exit 1
fi
gsm.orca > gsm.out 2> err &
The resulting path is written to stringfile.xyz0000 and can be used to find the transition state.
Frequencies
Identify the transition states for the pathfinder and GSM approaches and calculate frequencies for them at the GFN2-xTB (gas) level of theory.
xtb coord.xyz --hess > hess.out &
Have a look at the vibrational modes of the transition state in g98.out and vibspectrum with e.g. molden and check whether the correct TS was found and aligns with your chemical intuition and how both approaches compare.